Project Menhed Update #1: The Math Won (For Now)
The Silence (And The Grind)
I know, I’m late.
I haven’t posted in 36 days. I was planning on doing an update at Day 30, but honestly, I was too deep in a grind against OptKnock and my motivation was so down that I just couldn’t do it.
But I’m back! And I am ready to share the progress so far :D
The Learning Curve
For the past month, I spent a lot of time reading and self-teaching myself what Metabolic Engineering actually is. I learned from many resources and used AI to simplify the complex stuff.
It was long. It was tough. But I actually enjoyed it.
I read about 10+ papers (which is a lot to me! XD) to understand the tools and strategies researchers use. Some of the key papers that helped me:
- Designing Microbial Cell Factories for the Production of Chemicals
- Synergistic Production of Lycopene and Beta-Alanine Through Engineered Redox Balancing in Escherichia coli
- Production of Rainbow Colorants by Metabolically Engineered Escherichia coli (Really worth reading!)
- Metabolic Engineering of Escherichia coli for Natural Product Biosynthesis
The Work: Entering the Matrix
Once I learned the theory, I decided to get to work.
I looked up COBRApy and studied the documentation. I learned about Genome Scale Models (GEMs), Flux Balance Analysis (FBA), Flux Variability Analysis (FVA), and how to perform Knockouts for genes and reactions.
I dove deep into the E. coli core model to learn my way around reactions, metabolites, and genes. I learned how to find them, interpret what I see, how to add new ones and how to knock them out.
The Setup (In Silico)
Once I felt ready, I dove into the Lycopene Pathway.
I picked E. coli iML1515 as the GEM (since it is the most studied one) and jumped into the code. I introduced the Lycopene pathway, measured the theoretical maximum yield, and produced production envelopes.
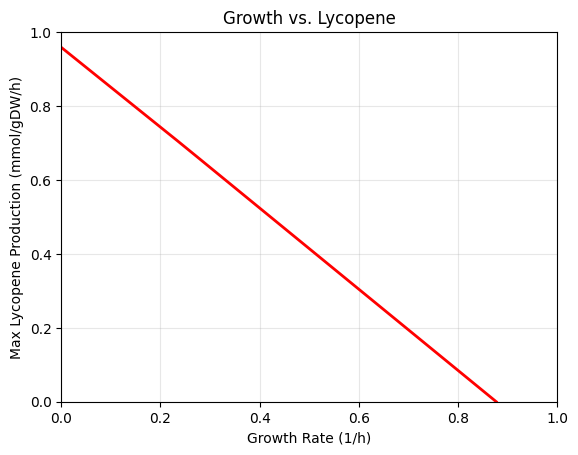
Growth vs Lycopene Production
You can check the code for this phase here: Project Menhed
The Crash (Why I Disappeared)
Then I decided to try and achieve Growth Coupling myself… and it didn’t go as expected XD.
First, I tried single reaction knockouts. Then doubles. None worked.
I tried to use OptKnock and it wasn’t easy at all. I endured through it, but just as I got it running, it would almost always crash or hang for hours.
I figured out the free GLPK solver just isn’t fit for this level of math. I looked into Gurobi, but unfortunately, my university isn’t on the list for a student license.
So I decided to go the hard way: Brute Force. I used Python loops to manually knockout reactions in Single, Double, and Triple combinations to figure it out.
It was a dead end.
Either there is no solution, or I am doing something totally wrong.
The Pivot: Round 2
So right now, I am going back in.
But this time, I will follow the literature. I will try to replicate processes others have done to reach coupling. If that works, yay.
If not, I’ll adopt a Two-Stage Fermentation scenario for now and dive deep into the fluxes to see what I can do to make the cell produce the Lycopene.
By the way, I found out that in my announcement post, I promised to use AlphaFold. Turns out, I probably won’t need it since the enzymes used in Lycopene Production are pretty well known already XD.
Work continues.
See you in the next log. (Hopefully sooner this time! XD)
All Updates will be available in the Progress Updates section of the announcement post