When Heart Disease Meets the Nervous System: Uncovering Immune Signatures in the Superior Cervical Ganglion

Preprint DOI: https://doi.org/10.1101/2025.05.18.654713
Ever wondered what happens to your nervous system when your heart is in trouble? It turns out, quite a lot! In this post, I’ll walk you through my recent reanalysis of RNAseq data from the superior cervical ganglion (SCG) in cardiac disease patients. What started as a curiosity-driven exploration revealed fascinating insights into neuroinflammation and immune signaling.
The SCG and its role in Cardiac Disease
The superior cervical ganglion sits quietly in your neck, controlling blood vessels, sweat glands, and–importantly for our story–aspects of heart function. Despite its critical role in cardiovascular regulation, we know surprisingly little about how this ganglion responds when the heart is diseased.
The Dataset: A Second Look at Valuable Data
For this study, I revisited human bulk RNAseq data originally published by Ziegler et al. (2023), available under ENA accession number PRJNA967653. The dataset includes SCG samples from:
- 3 patients with cardiac disease
- 3 healthy controls
While the original authors focused primarily on single-cell RNAseq findings, I saw an opportunity to extract additional value from the human bulk RNAseq datasets, a perfect example of how reanalyzing existing data can yield new discoveries without additional experimental costs.
Methods: Data Processing Pipeline
I implemented a comprehensive analysis pipeline that included:
- Quality Control: Using FastQC via Galaxy platform
- Pseudoalignment: Employing kallisto against the human GRCh38 transcriptome
- Differential Expression: Analyzing with DESeq2, filtering for adjusted p-value < 0.05 and abs(log2FC) ≥ 1
- Functional Enrichment: Applying GO, KEGG, and GSEA analyses using clusterProfiler and fgsea
All code and analysis scripts are available on GitHub for complete reproducibility: GitHub Repository
Analysis Results
Sample Relationships Reveal Heterogeneity
One of the first surprises came from the Principal Component Analysis (PCA):
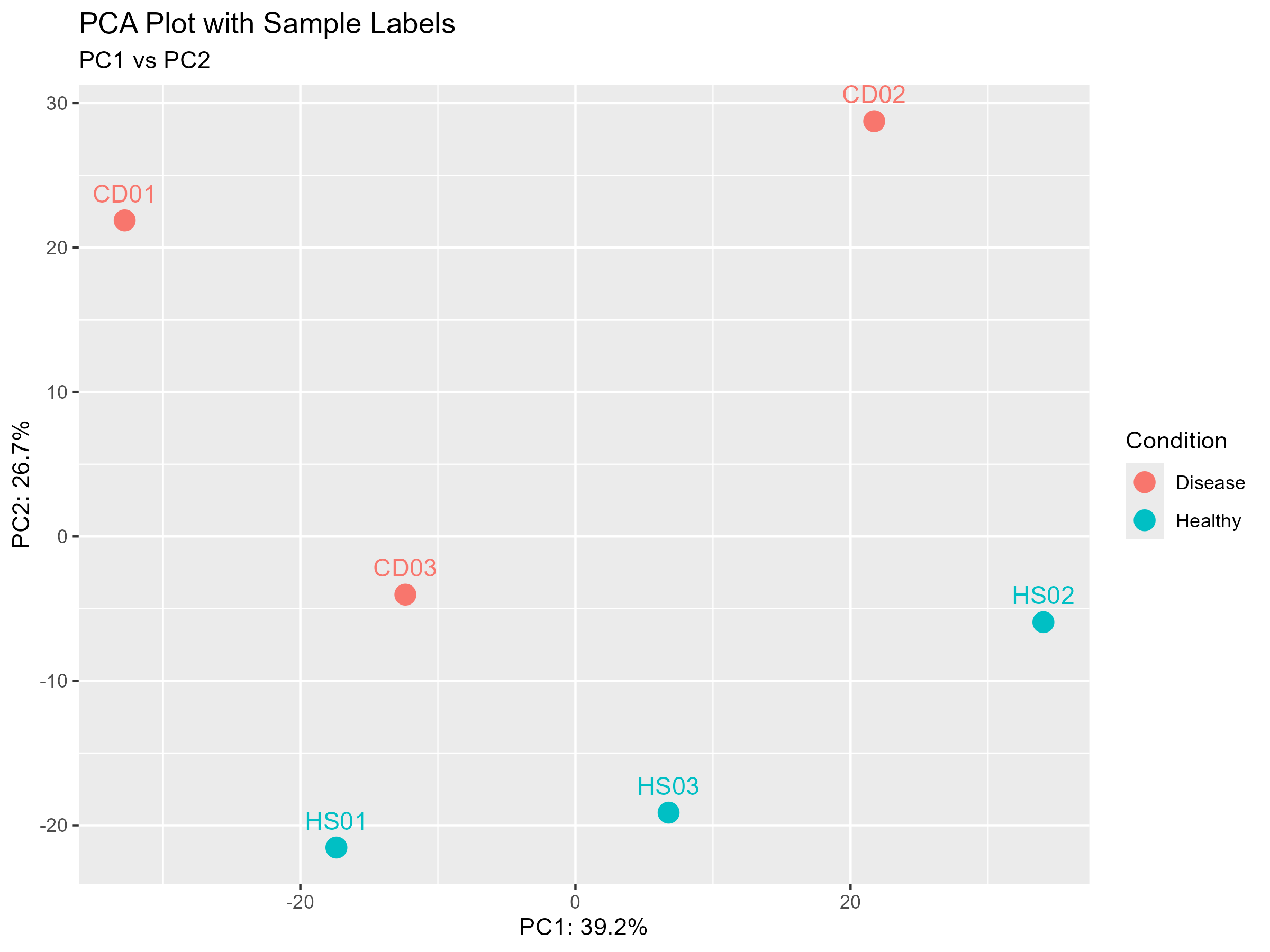
Rather than a clean separation between healthy and disease samples, I observed two distinct clusters that each contained both healthy and disease samples. This can suggest considerable biological heterogeneity and early or subclinical disease states.
The sample-to-sample distance heatmap further confirmed this pattern:
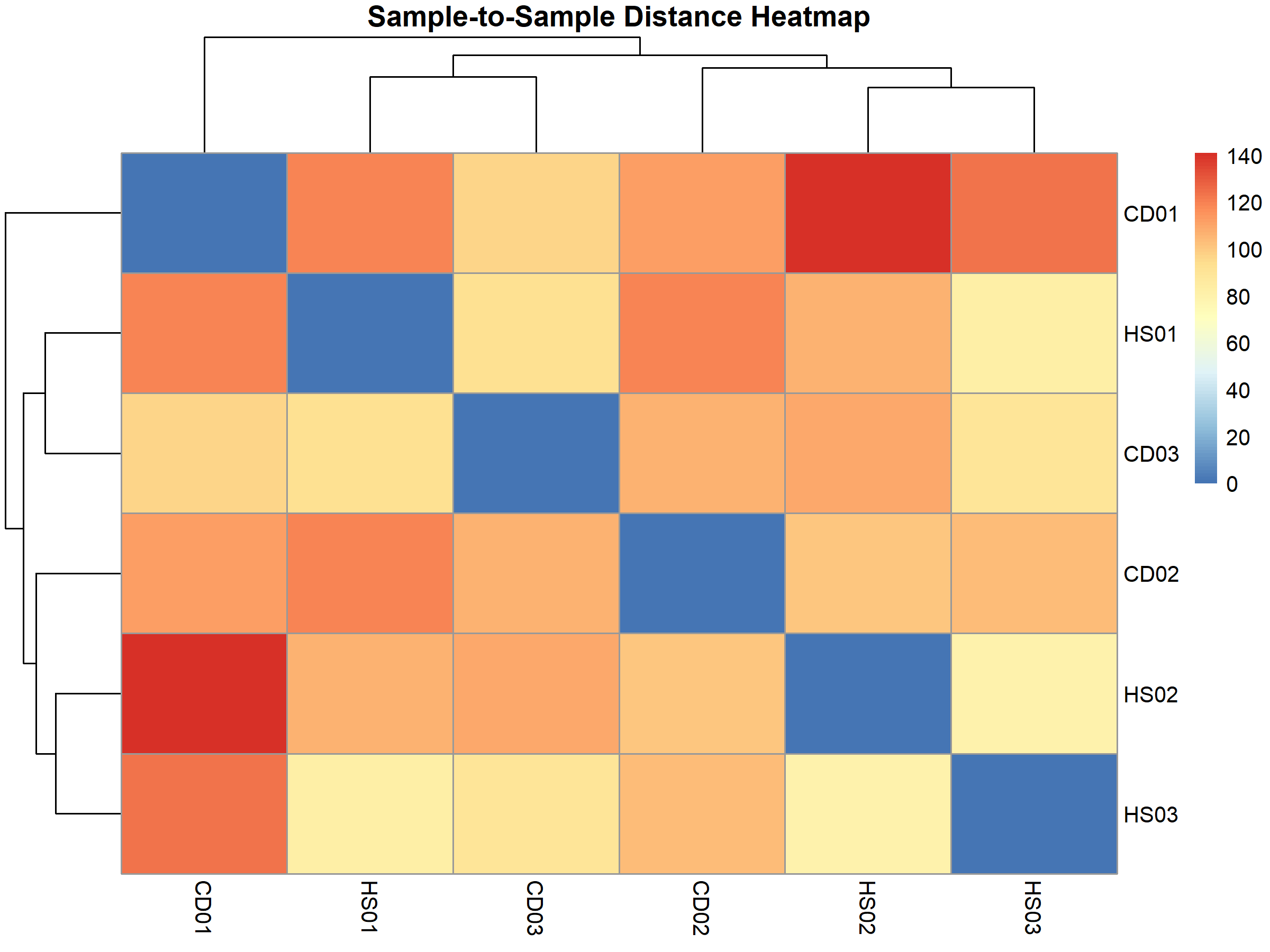
This clustering pattern persisted even after correcting for unwanted variation using Surrogate Variable Analysis (SVA), suggesting it reflects genuine biological differences rather than technical artifacts.
Differential Expression Analysis
The differential expression analysis identified 771 significantly altered genes between cardiac disease and healthy SCG samples:
- 399 upregulated genes
- 372 downregulated genes
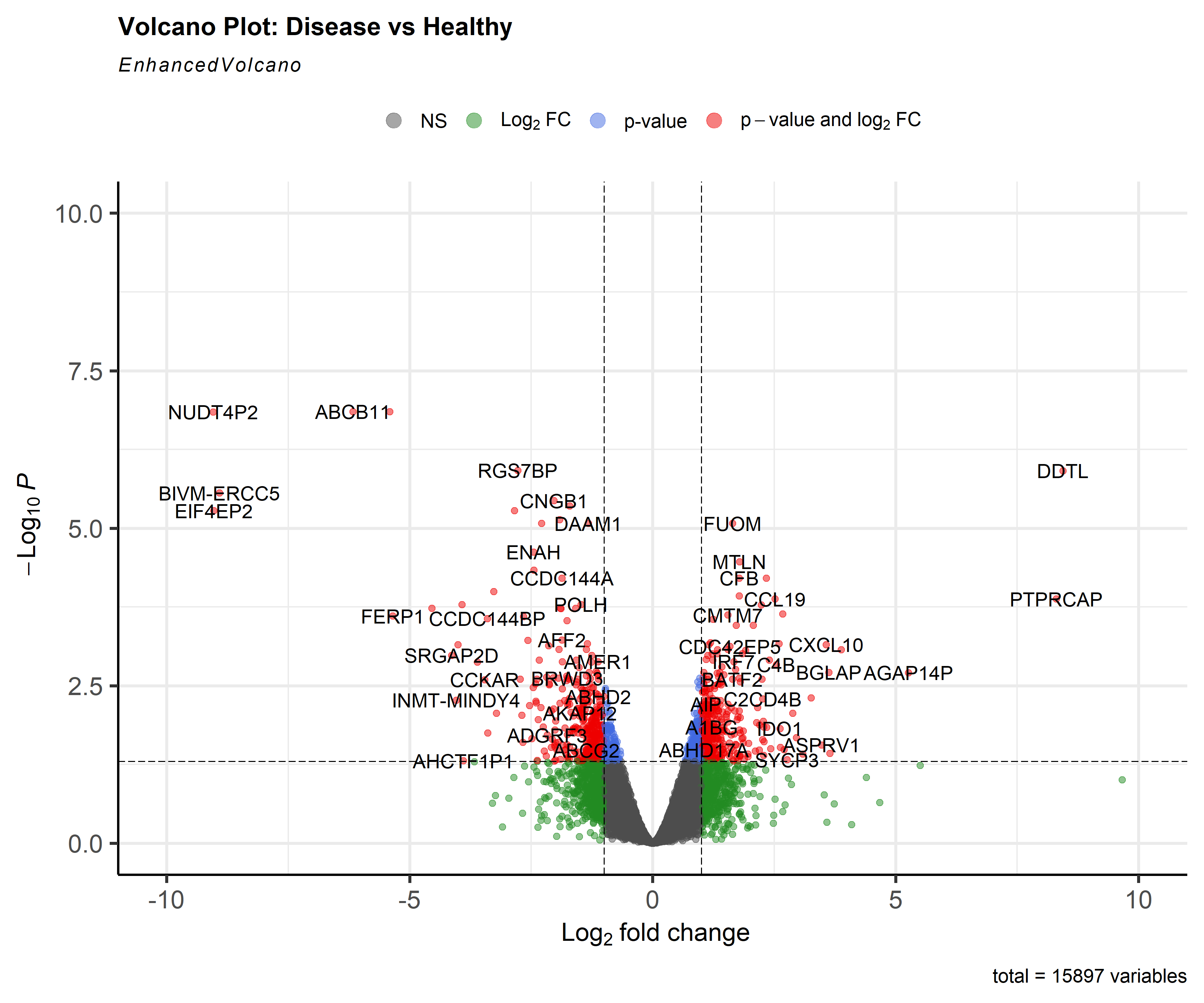
Among the highest upregulated genes were DDTL and PTPRCAP. Meanwhile, NUDT4P2, ABCB11, and BIVM-ERCC5 showed strong downregulation.
When examining the expression patterns of inflammatory markers across samples, clear differences emerged:
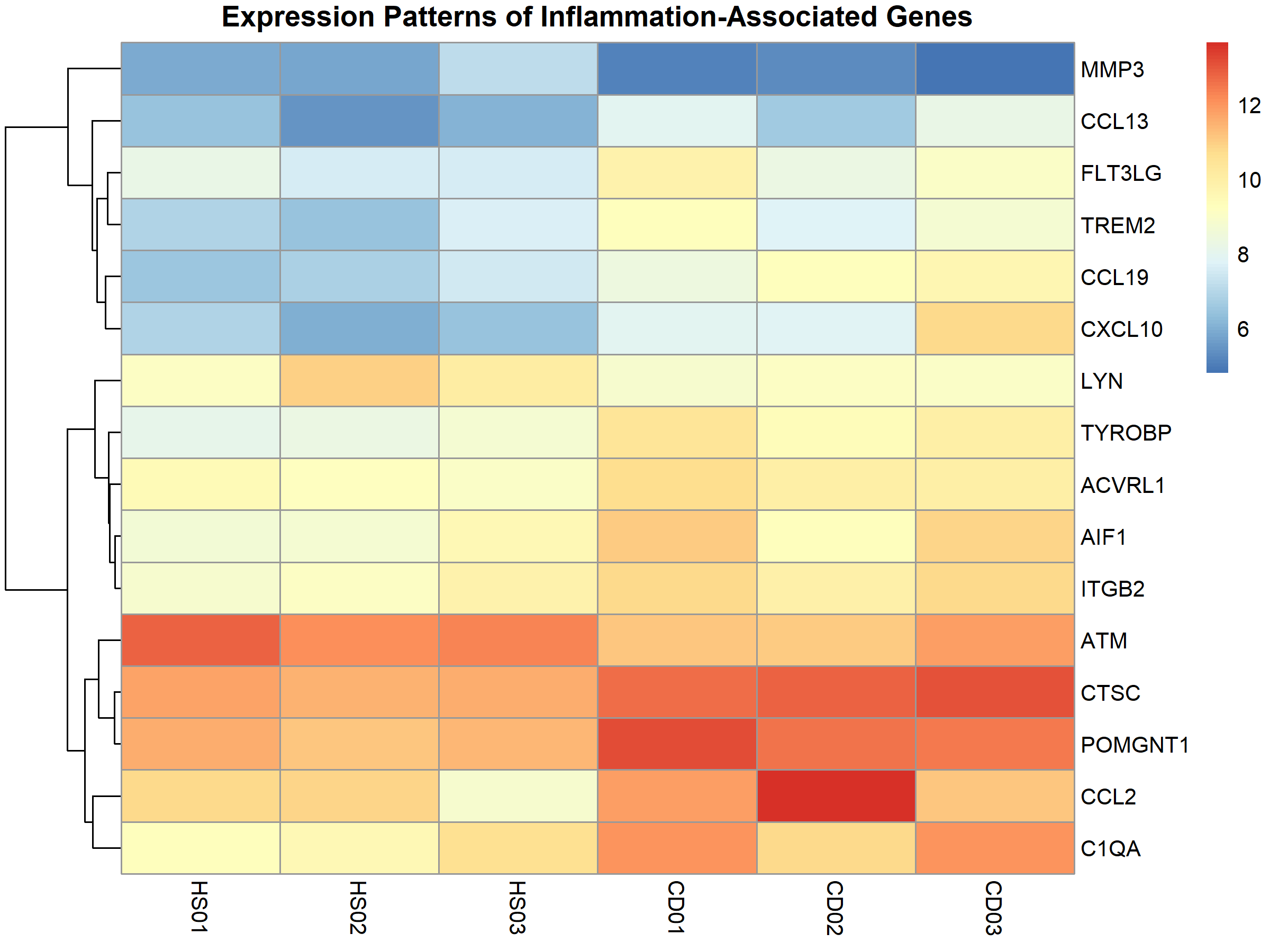
Similarly, immunity-related genes showed distinct expression patterns between disease and healthy samples:
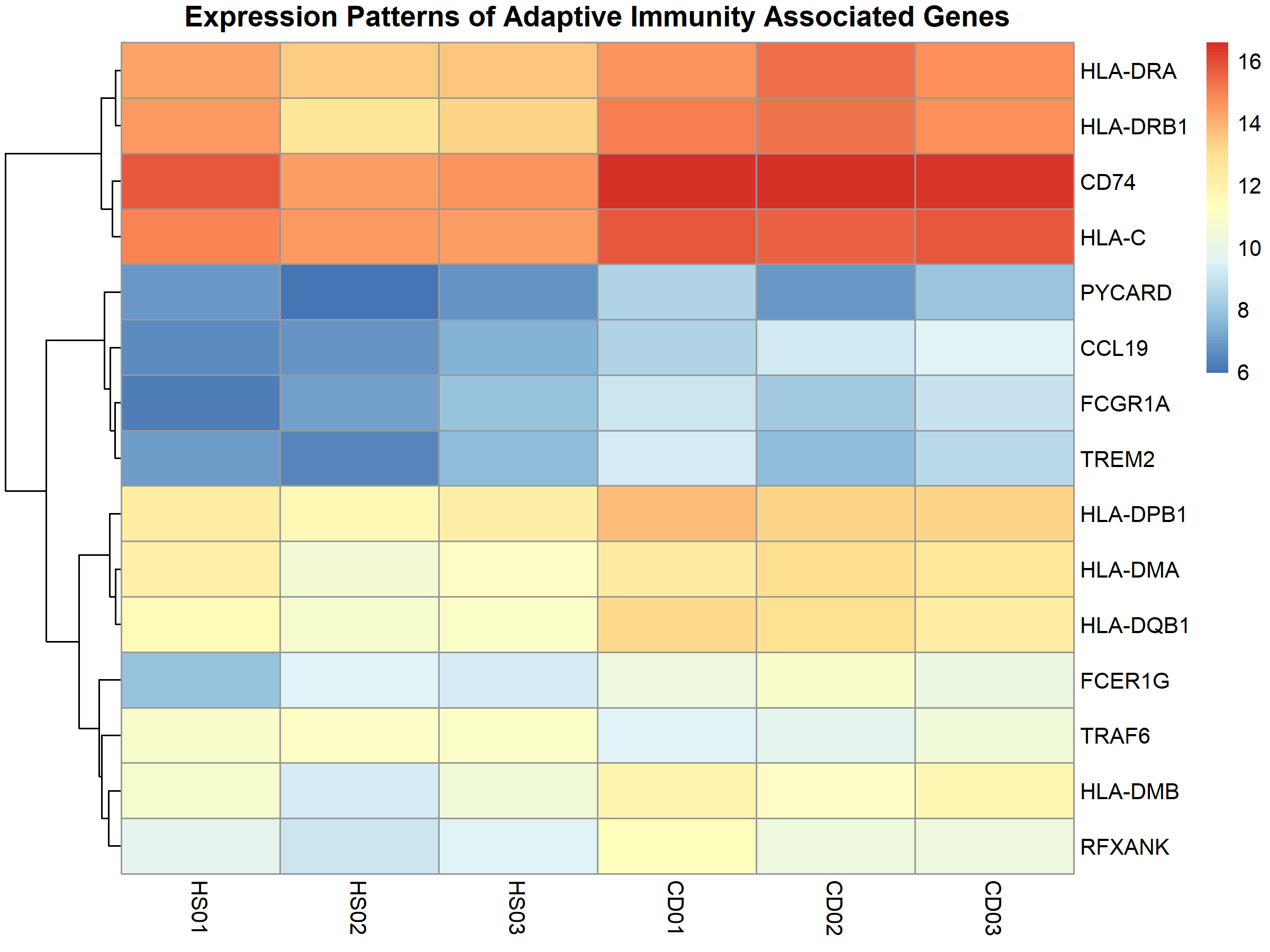
Functional Enrichment & GSEA Analyses: Connecting the Dots
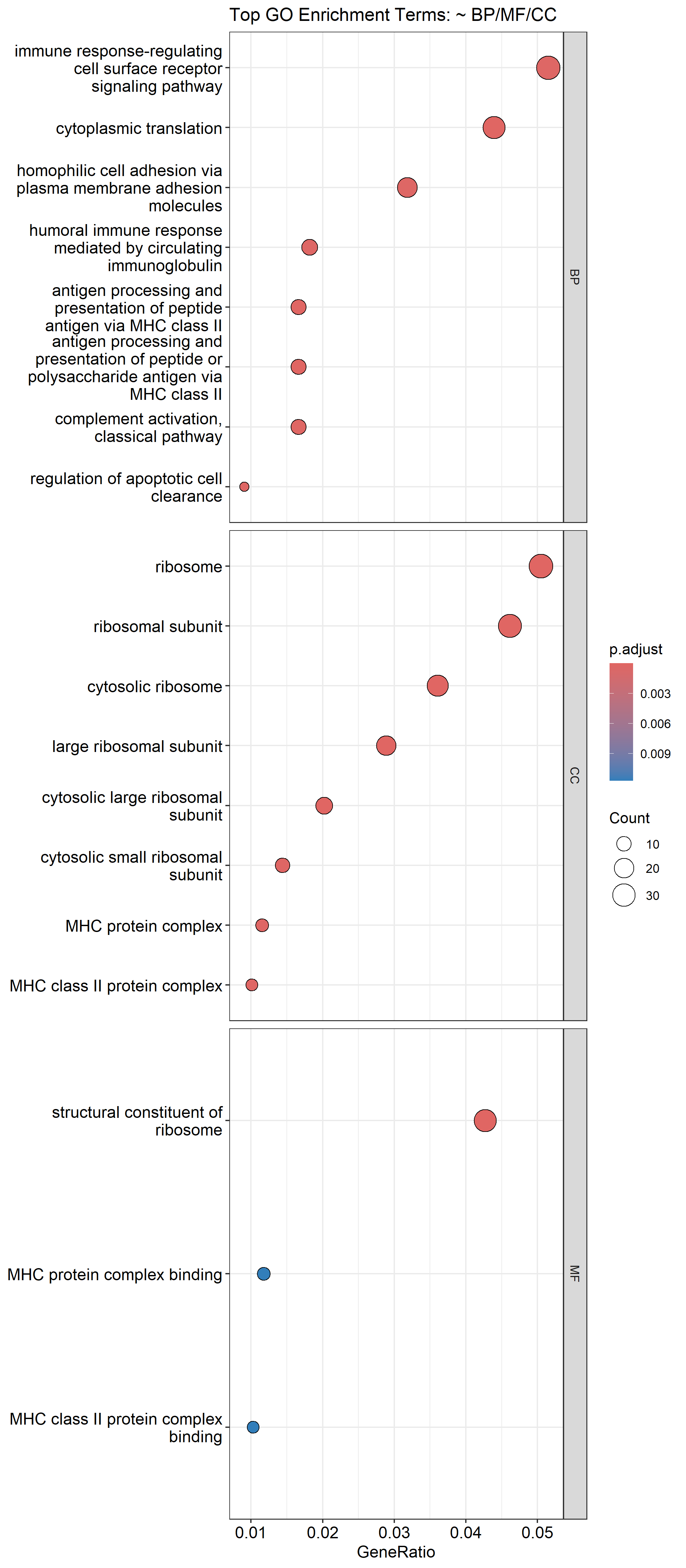
The Gene Ontology (GO) enrichment analysis revealed 148 significantly enriched terms across biological processes, cellular components, and molecular functions. The top biological processes included:
- Immune response-regulating cell surface receptor signaling
- Cytoplasmic translation
- Antigen processing and presentation via MHC class II
KEGG pathway analysis identified 19 enriched pathways, with several related to immune function and inflammation:
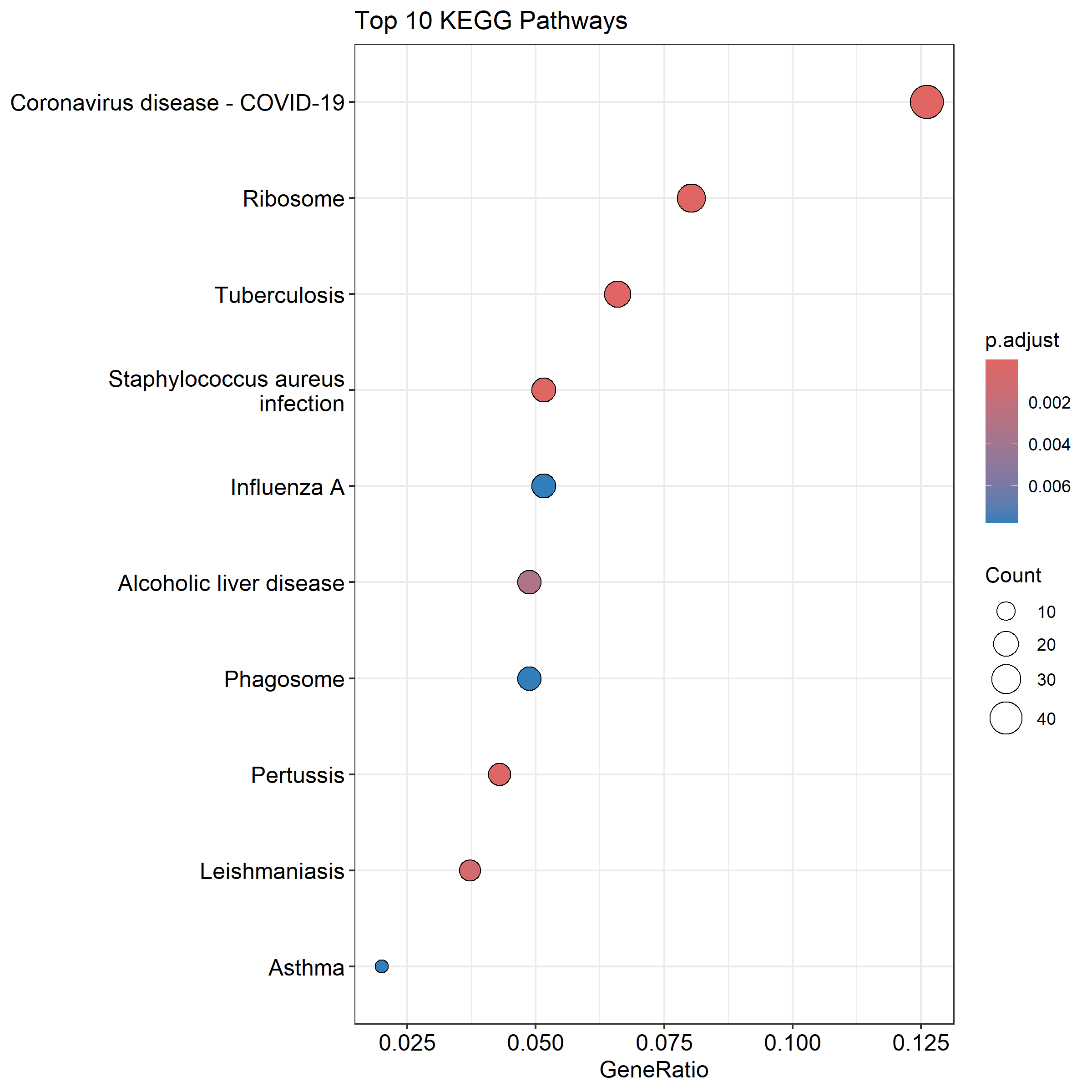
Interestingly, pathways related to infectious diseases like “Coronavirus disease – COVID-19” and “Staphylococcus aureus infection” were enriched. This doesn’t mean the patients had these infections, rather, it reflects shared immune and inflammatory mechanisms between cardiac disease and infectious responses.
GSEA identified 40 enriched gene sets, with particularly strong enrichment in cytokine-cytokine receptor interactions And antigen processing and presentation further supporting the idea of an active neuroimmune inflammatory response in the SCG during cardiac disease.
The Big Picture: Neuroinflammation, Immunity, and Metabolism
Putting all these findings together reveals three major themes in the SCG’s response to cardiac disease:
1. Neuroinflammation Takes Hold
The data shows clear evidence of inflammatory processes within the SCG, including:
- Upregulation of inflammatory chemokines
- Increased expression of macrophage activation markers
- Enhanced complement system component expression
This suggests that the SCG doesn’t just passively respond to cardiac disease, it actively participates in inflammatory processes that may influence disease progression.
2. Adaptive Immunity Enters the Scene
Perhaps most surprisingly, I found significant enrichment of adaptive immune processes, marked by:
- Increased expression of multiple MHC class I and II genes
- Enhanced antigen presentation activity
- Upregulation of T-cell signaling components
This raises questions about immune cell recruitment to the SCG during cardiac disease and potential neuroimmune interactions that haven’t been previously appreciated.
3. Metabolic Reprogramming Supports Immune Function
The third major theme involves metabolic shifts within the SCG:
- Increased expression of mitochondrial ATP synthesis genes
- Enrichment of oxidative phosphorylation pathways
- Enhanced purine metabolism
These metabolic changes likely support the increased energy demands of inflammatory processes and suggest a coordinated immuno-metabolic response within the ganglion.
Limitations and Future Directions
While these findings are exciting, several limitations should be acknowledged:
- Small sample size: With only three samples per group, statistical power is limited
- Sample heterogeneity: The unexpected clustering patterns suggest biological variability that warrants further investigation
- Lack of protein-level validation: Transcriptomic changes don’t always translate to functional protein differences
Future studies should aim to validate these findings in larger cohorts & Investigate protein-level changes in the SCG during cardiac disease
Conclusion: The Value of Data Reanalysis
These findings suggest that the SCG may serve as an active immuno-metabolic hub during cardiac disease, potentially influencing both neural control of the heart and systemic inflammatory responses.
Could targeting neuroinflammation in the SCG provide a novel therapeutic approach for cardiac patients? Could the SCG serve as a biomarker site for disease progression?
This study demonstrates the tremendous value of revisiting existing transcriptomic datasets with new analytical approaches. By taking a second look at the SCG bulk RNAseq data, I’ve uncovered evidence for neuroinflammation, adaptive immunity, and metabolic reprogramming that adds new dimensions to our understanding of the heart-brain connection in disease.
Access the Full Study
This reanalysis is available as a preprint on bioRxiv:
https://doi.org/10.1101/2025.05.18.654713
All code, analysis scripts, and supplementary files can be found on GitHub:
https://github.com/DjoserGenomics/SCG-cardiac-disease-reanalysis
Citation
If you reference this work, please cite:
Nourelden Rihan. Transcriptomic Signatures of Neuroinflammation and Adaptive Immunity in the Human SCG During Cardiac Disease: A Bulk RNAseq Reanalysis. bioRxiv 2025.
DOI: 10.1101/2025.05.18.654713