Field Note 13: Dispatch - HTGAA Week 6 Homework

1. What are some components in the Phusion High-Fidelity PCR Master Mix and what is their purpose?
- Phusion DNA Polymerases: this builds the new copied DNA strand
- dNTPs (A, T, C, G): these are the building blocks of DNA
- Buffer & MgCl2: these help the DNA polymerases work
2. What are some factors that determine primer annealing temperature during PCR?
The two main factors are the Primer Length and GC Content. These determine the Melting Temperature (Tm) - the temperature at which half the primers fall off the DNA.
3. There are two methods in this protocol that create linear fragments of DNA: PCR, and restriction enzyme digest. Compare and contrast these two methods, both in terms of protocol as well as when one may be preferable to use over the other.
Restriction enzymes can be useful as their cuts are precise and does not induce mutations but it depends on the availability of restriction sites and if they are in areas far away from important segments in the DNA or plasmid like the Origin of Replication (ORI) and other segments inside target genes or selection markers, Also Using restriction enzymes mostly would leave sticky edges that need to be dealt with using primers or overhangs, PCR can overcome this as it doesn’t need to make a cut, a primer can be designed to stick to a certain part and skip another allowing for a seamless copying but it can have erros and induce mutations especially with large segments of DNA
4. Why does the PvuII digest require CutSmart buffer?
Restriction enzymes like PvuII are proteins that require very specific chemical conditions to function efficiently. The CutSmart buffer provides the optimal pH, salt concentration, and essential cofactors that PvuII needs to cut DNA effectively. Else the enzyme would be inactive or cut at the wrong sites.
5. How can you ensure that the DNA sequences that you have digested and PCR-ed will be appropriate for Gibson cloning?
This is ensureed during the Design phase by designing specific PCR primers. The primers must be designed to add homologous overlaps (20–40 bp) to the ends of the PCR fragment that perfectly match the ends of the linearized plasmid. That way the Gibson enzymes can successfully chew back and anneal them together.
6. How does the plasmid DNA enter the E. coli cells during transformation?
The DNA enters through a multiple processes including Heat Shock or Electroporation. First, the chemically competent cells (treated with Calcium) allow the negatively charged DNA to stick to the bacterial cell wall. Then, a rapid increase in temperature (usually to 42°C for 45 seconds) creates a thermal imbalance and transient pores in the cell membrane, which effectively sucks the plasmid DNA from the outside into the cytoplasm. Or Electroporation where a high-voltage pulse is used to create temporary pores in the cell membrane, allowing the DNA to enter the cells.
**7. Describe another assembly method in detail (such as Golden Gate Assembly) 5 - 7 sentences w/ diagrams (either handmade or online). **Model this assembly method with Benchling or a similar tool!**
Golden Gate Assembly is a powerful, modular cloning method that relies on Type IIS restriction enzymes (such as BsaI or BsmBI), which have the unique ability to cut DNA outside of their specific recognition sequence. This allows us to design custom 4-base pair overhangs on the ends of DNA fragments that act as “barcodes” ensuring that multiple parts (like a promoter, RBS, and gene) assemble in the exact correct order. Because the enzyme cuts the recognition site off the fragment, the digestion and ligation can happen simultaneously in a single “one-pot” reaction. If the parts ligate correctly, the recognition site is lost, making the reaction irreversible and efficient. This results in a completely scarless assembly, making it ideal for building complex, multi-gene circuits.
Used Gemini’s Nano Banana Pro to produce this infographic :D
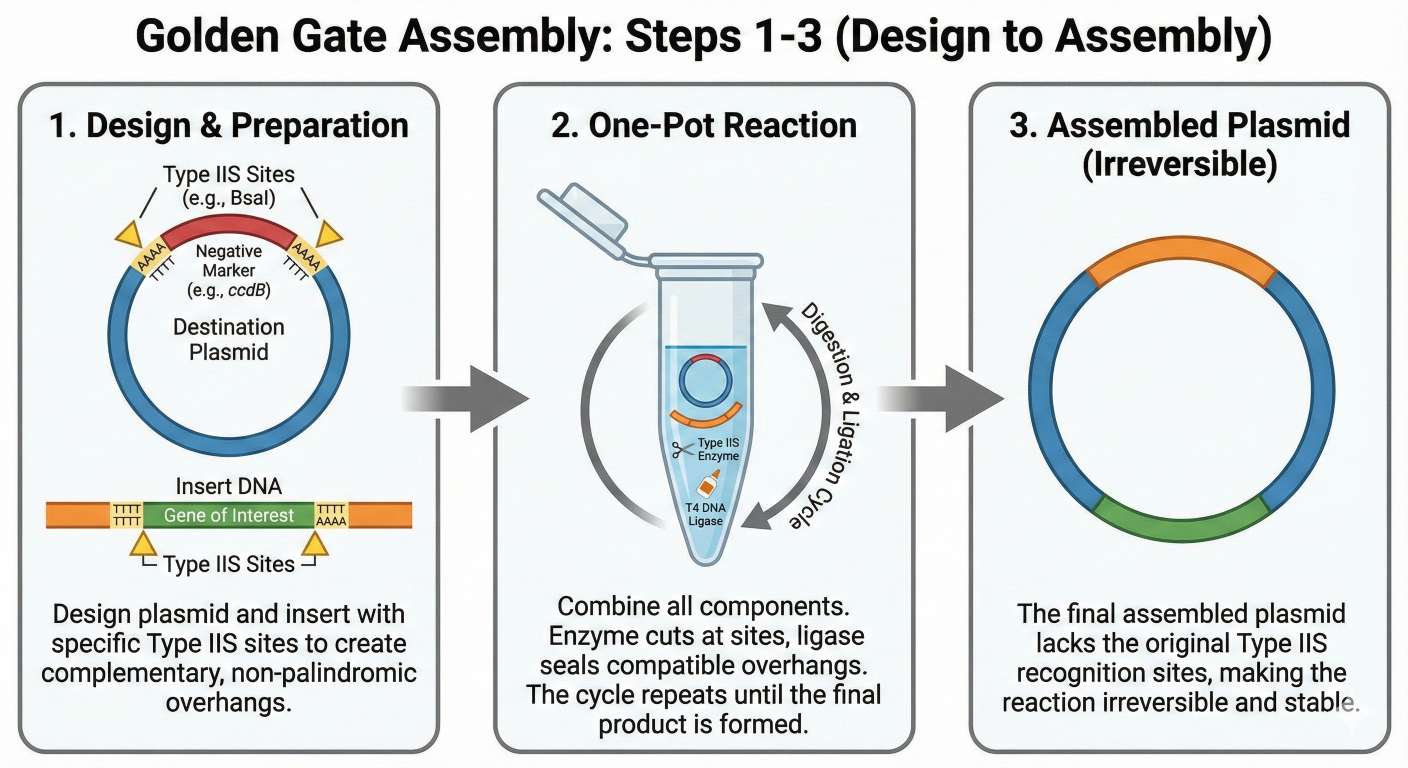
Golden Gate Assembly Infographic
Check out Other HTGAA Submissions from Here