Field Note 12: Dispatch - HTGAA Week 5 Homework

Part A (From Pranam)
I generated Two Binder peptides for the SOD1 (A4V mutated) Protein in additon to the literature binder peptide FLYRWLPSRRGG that was provided in the homework assignment.
| Number | Binder | Pseudo Perplexity |
|---|---|---|
| 0 | WRVYVVAVALKE | 21.914981 |
| 1 | WRYYVAAAALGE | 11.314945 |
| 2 | FLYRWLPSRRGG | n/a |
I then extracted the ipTM scores from the log files generated by AlphaFold-Multimer for each seed of the three binder peptides complexed with the SOD1 (A4V mutated) Protein and plotted them into a bar chart using matplotlib.
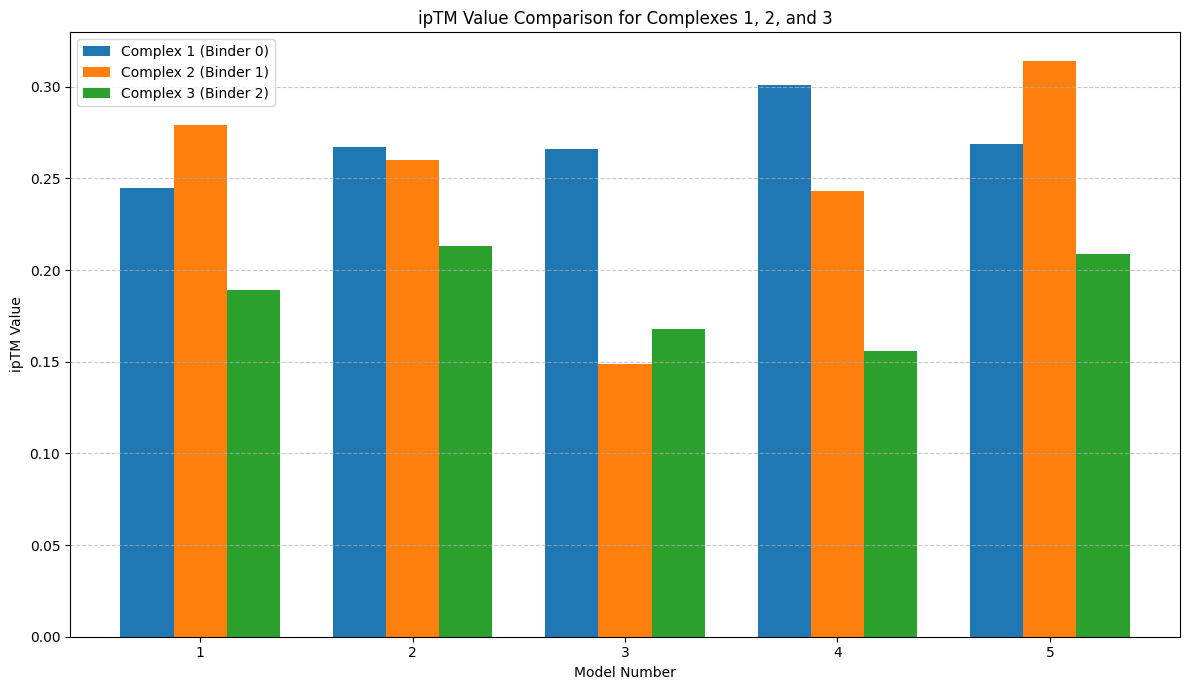
Bar Chart of ipTM Scores for Binder Peptides Complexed with SOD1 (A4V mutated) Protein
The bar chart compares the ipTM values for five models/seeds across three different Binders (complexes). Binder 2 has some of the highest ipTM values, peaking at 0.314 for model 5. Binder 1 also shows high ipTM values, with its highest at 0.301 for model 4. Binder 3 consistently has the lowest ipTM values across all models, with its highest value being 0.213 for model 2. Overall, Binder 2 appears to have the best performance based on ipTM values, while Binder 3 performs the worst among them.
Part B (Final Project: L-Protein Mutants)
Decided to go with OPTION 2: Follow the below pipeline to engineer the L-protein!, The reason i didn’t go with my Armored Phage Proposal From Last Week’s Homework Assignment, is that after going through it again, i believe it still lacks more validation or “Success Metrics” and more thermostability-related tools to back it up. So for now i will be focusing on engineering the L-protein.
This heatmap was generated using this notebook
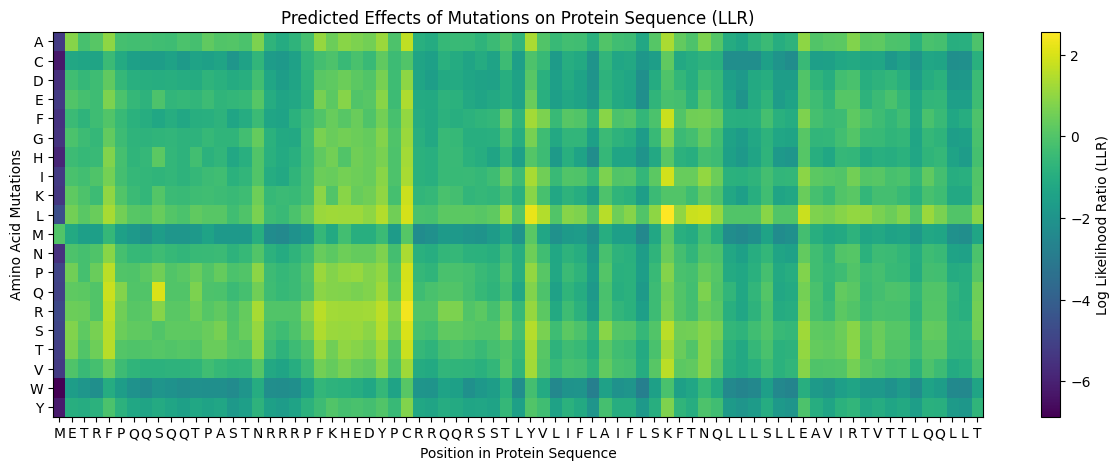
A Heatmap Showing the Predicted Effects of Mutations on the L Protein Sequence
We need to identify mutations that have a possible enhancing effect on the lysis activity of the L Protein, mainly we need to select mutations that:
- Have a positive
LLRscore, indicating this is a favorable mutation. - Should not be in the conserved regions of the protein.
- Maintains the Lysis activity of the L Protein.
- Does not disrupt the coat and replication Proteins of the MS2 Phage.
To identify the conservative pBlast and Clustal Omega were used to identify the conserved regions of the L Protein. Here is a link to the Clustal Omega job that was used to identify the conserved regions of the L Protein.
To Better Visualize the conserved regions of the L Protein, I imported the L Protein sequence into Benchling and annotated the conserved regions based on the Clustal Omega results. I also annotated the Soluble Domain (Amino Acids 1-40) and the Transmembrane Domain (Amino Acids 41-75).

L Protein Sequence with Annotated Conserved Regions in Benchling (Gray Tracks are the conserved Regions)
Interesting Observation: It seems that all the conserved regions are concentrated in the soluble domain, the one responsible for interacting with the host’s chaperone DnaJ, while the transmembrane domain seems to be less conserved.
I selected the following mutations as potential candidates for enhancing the lysis activity of the L Protein:
| Mutation Codename | Amino Acid Position | Amino Acid Change | LLR Score | Region | Overlaps with Coat/Rep protein? |
|---|---|---|---|---|---|
| K50L | 50 | K->L | 2.56146419048309 | Transmembrane | Yes |
| Y39L | 39 | Y->L | 2.24177801609039 | Soluble | Yes |
| S09Q | 9 | S->Q | 2.01432251930236 | Soluble | Yes |
| K50I | 50 | K->I | 1.92879796028137 | Transmembrane | Yes |
| Y27R | 27 | Y->R | 1.62805962562561 | Soluble | No |
Check out Other HTGAA Submissions from Here