Armored Phage: A Thermostable MS2 Phage Proposal

A Proposal to Engineer a Thermostable MS2 Bacteriophage Using Protein Design Techniques.
This Proposal is part of How To Grow Almost Anything (HTGAA) Course Homework for Week 4 (Protein Design), Check the Week 4 Homework for more details.
Project Goal
The Goal of this project is to engineer a thermostable variant of the MS2 bacteriophage Capsid Protein through reengineering the Capsid Protein to be more resistant to high temperatures. This will help create a robust phage, which is critical for applications like phage therapy.
Our Proposed Pipeline and Tools
Our pipeline is 100% computational and leverages state-of-the-art protein design and structure prediction tools:
-
Analyze the MS2 Capsid Protein Structure using tools like PyMOL or ChimeraX (
PDB ID: 2MS2):- Understand how the individual capsid protein monomers lock together to form the icosahedral shell.
- Identify key structural features and regions that contribute to thermal stability.
- Map out potential weakness sites that a mutation or an edit could enhance its thermal stability.
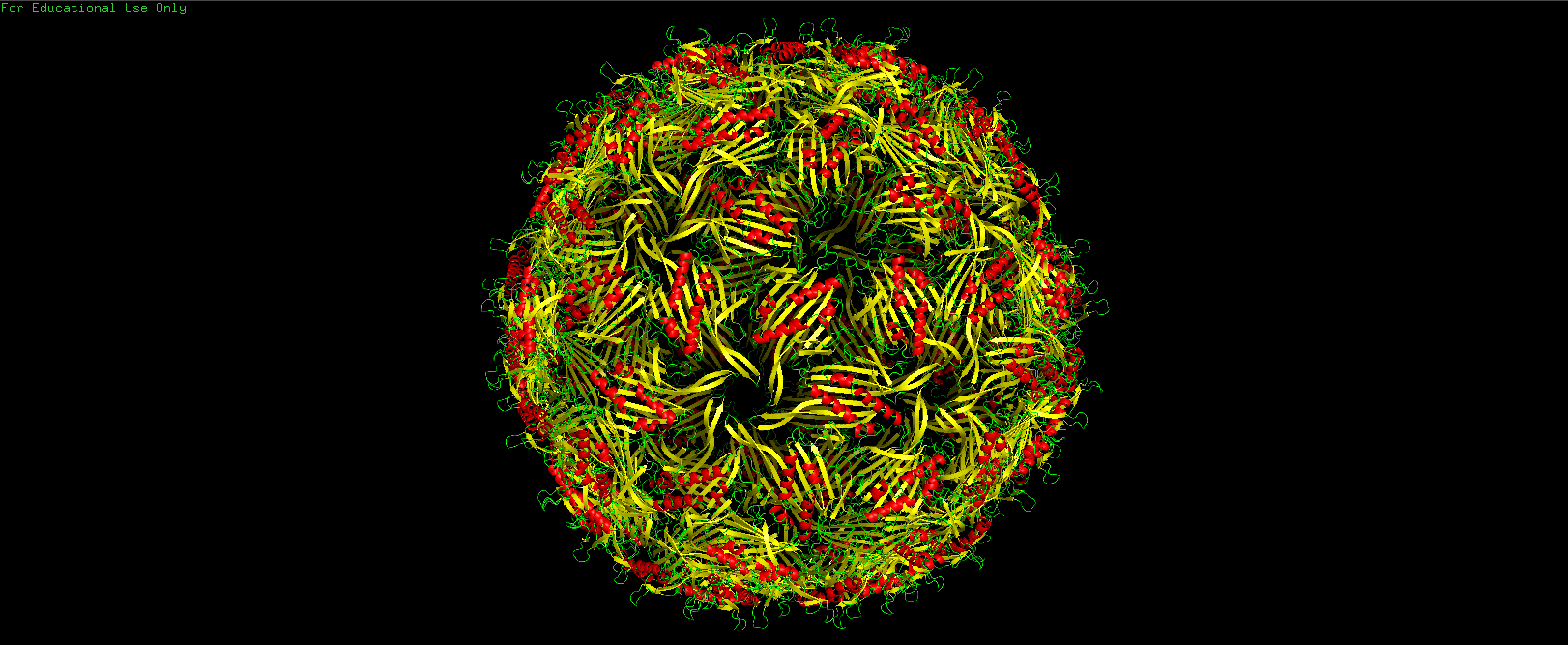
PyMOL Visualization showing the full icosahedral 'shell' of the MS2 bacteriophage (PDB: 2MS2).
-
Use AlphaFold to Identify targets in the structure of a single Capsid Protein Monomer:
- The Target is to identify flexible regions or loops that are prone to thermal denaturation.
- Usually these regions can be highlighted by low confidence scores in AlphaFold predictions.

PyMOL Visualization showing a single 'monomer' or 'building block' of the MS2 capsid protein. (Yellow is Beta Sheets, Red is Alpha helices, Green is in between loops)
-
Use ProteinMPNN to try and design mutations in the Capsid Protein:
- Stable Regions would be masked to keep intact.
- Only flexible regions would be targeted for mutations.
-
Predict and verify the structure of the mutated Capsid Protein Monomer using AlphaFold:
- Ensure that the overall fold of the protein is maintained.
- Check if the mutations introduced have improved the stability of the protein.
- Look for increased hydrogen bonding, salt bridges, and hydrophobic interactions.
Why These Tools?
-
PyMOL/ChimeraX: These are powerful molecular visualization tools that allow us to view and analyze protein structures, making it easier to identify key features related to stability.
-
AlphaFold: It provides highly accurate protein structure predictions, which is essential for identifying flexible regions and understanding how mutations may affect the protein’s 3D folding.
-
ProteinMPNN: It designs a new protein sequence based on 3D structural information, making it ideal for suggesting mutations that could enhance thermal stability and allows for masking stable regions to preserve essential structural features.
Potential Pitfalls
-
Function Loss: By making the capsid protein more rigid and heat stable, we may unintentionally interfere with its ability to infect host bacteria or dirupt its assembly into a functional phage capsid especially that it has a quaternary structure or .
-
Prediction Erros: Our entire pipeline relies on computational predictions, which may not always accurately reflect real behaviour. Experimental validation would be necessary to confirm the effectiveness of the designed mutations.
-
Limited Mutation Space: The capsid protein may have limited sites that can be mutated without disrupting its structure, function, or folding which can limit our design options.
Pipeline Schematic
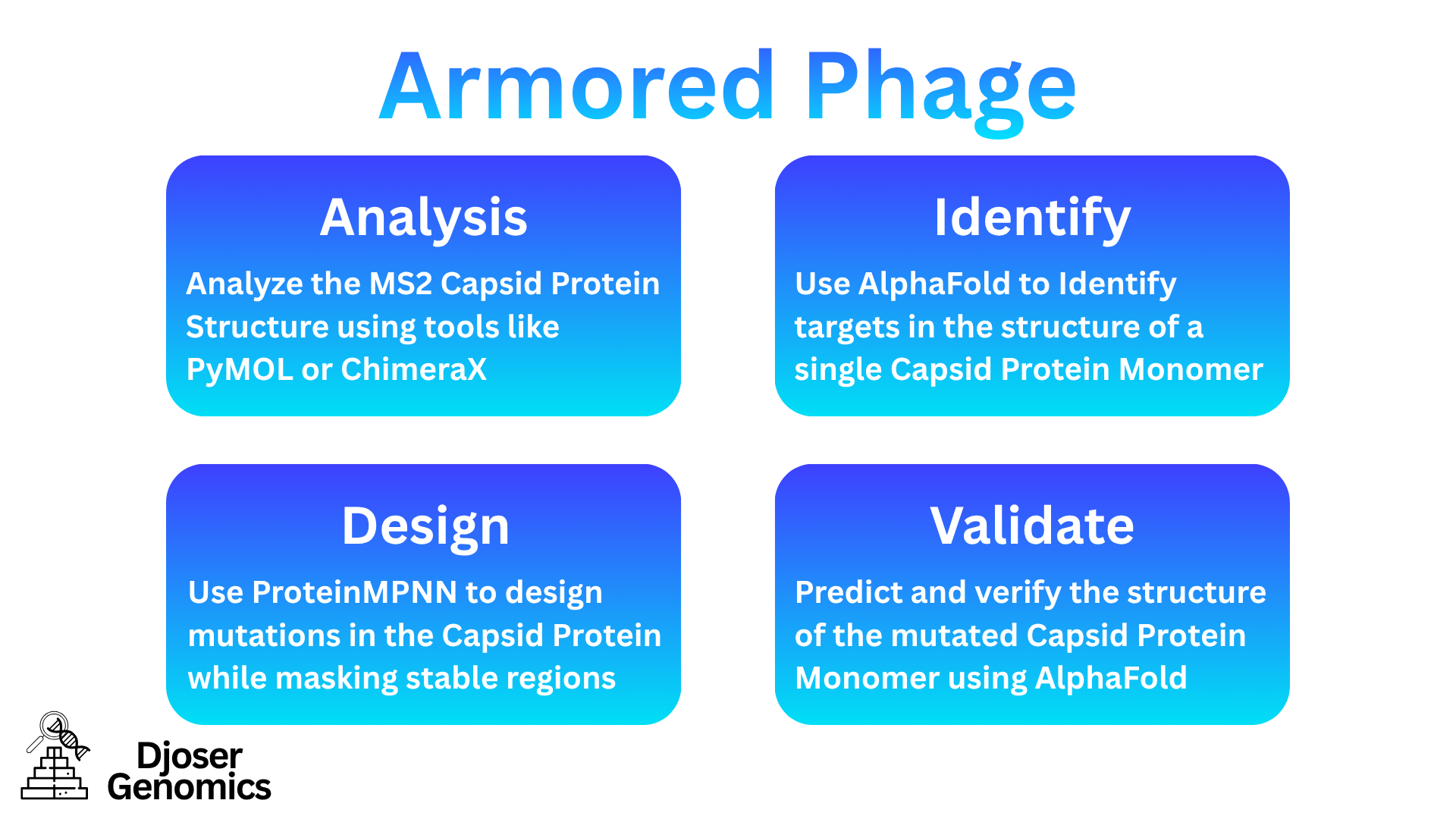
A Schematic Showing a Brief Overview of the Proposed Pipeline